
Scatterplot with marginal distributions and statistical results
Source:R/ggscatterstats.R
ggscatterstats.RdScatterplots from {ggplot2} combined with marginal distributions plots
with statistical details.
Usage
ggscatterstats(
data,
x,
y,
type = "parametric",
conf.level = 0.95,
bf.prior = 0.707,
bf.message = TRUE,
tr = 0.2,
digits = 2L,
results.subtitle = TRUE,
label.var = NULL,
label.expression = NULL,
marginal = TRUE,
point.args = list(size = 3, alpha = 0.4, stroke = 0),
point.width.jitter = 0,
point.height.jitter = 0,
point.label.args = list(size = 3, max.overlaps = 1e+06),
smooth.line.args = list(linewidth = 1.5, color = "blue", method = "lm", formula = y ~
x),
xsidehistogram.args = list(fill = "#009E73", color = "black", na.rm = TRUE),
ysidehistogram.args = list(fill = "#D55E00", color = "black", na.rm = TRUE),
xlab = NULL,
ylab = NULL,
title = NULL,
subtitle = NULL,
caption = NULL,
ggtheme = ggstatsplot::theme_ggstatsplot(),
ggplot.component = NULL,
...
)Arguments
- data
A data frame (or a tibble) from which variables specified are to be taken. Other data types (e.g., matrix,table, array, etc.) will not be accepted. Additionally, grouped data frames from
{dplyr}should be ungrouped before they are entered asdata.- x
The column in
datacontaining the explanatory variable to be plotted on thex-axis.- y
The column in
datacontaining the response (outcome) variable to be plotted on they-axis.- type
A character specifying the type of statistical approach:
"parametric""nonparametric""robust""bayes"
You can specify just the initial letter.
- conf.level
Scalar between
0and1(default:95%confidence/credible intervals,0.95). IfNULL, no confidence intervals will be computed.- bf.prior
A number between
0.5and2(default0.707), the prior width to use in calculating Bayes factors and posterior estimates. In addition to numeric arguments, several named values are also recognized:"medium","wide", and"ultrawide", corresponding to r scale values of1/2,sqrt(2)/2, and1, respectively. In case of an ANOVA, this value corresponds to scale for fixed effects.- bf.message
Logical that decides whether to display Bayes Factor in favor of the null hypothesis. This argument is relevant only for parametric test (Default:
TRUE).- tr
Trim level for the mean when carrying out
robusttests. In case of an error, try reducing the value oftr, which is by default set to0.2. Lowering the value might help.- digits
Number of digits for rounding or significant figures. May also be
"signif"to return significant figures or"scientific"to return scientific notation. Control the number of digits by adding the value as suffix, e.g.digits = "scientific4"to have scientific notation with 4 decimal places, ordigits = "signif5"for 5 significant figures (see alsosignif()).- results.subtitle
Decides whether the results of statistical tests are to be displayed as a subtitle (Default:
TRUE). If set toFALSE, only the plot will be returned.- label.var
Variable to use for points labels entered as a symbol (e.g.
var1).- label.expression
An expression evaluating to a logical vector that determines the subset of data points to label (e.g.
y < 4 & z < 20). While using this argument withpurrr::pmap(), you will have to provide a quoted expression (e.g.quote(y < 4 & z < 20)).- marginal
Decides whether marginal distributions will be plotted on axes using
{ggside}functions. The default isTRUE. The package{ggside}must already be installed by the user.- point.args
A list of additional aesthetic arguments to be passed to the
ggplot2::geom_point().- point.width.jitter, point.height.jitter
Degree of jitter in
xandydirection, respectively. Defaults to0(0%) of the resolution of the data. Note that the jitter should not be specified in thepoint.argsbecause this information will be passed to two differentgeoms: one displaying the points and the other displaying the *labels for these points.- point.label.args
A list of additional aesthetic arguments to be passed to
ggrepel::geom_label_repel()geom used to display the labels.- smooth.line.args
A list of additional aesthetic arguments to be passed to
geom_smoothgeom used to display the regression line.- xsidehistogram.args, ysidehistogram.args
A list of arguments passed to respective
geom_s from the{ggside}package to change the marginal distribution histograms plots.- xlab
Label for
xaxis variable. IfNULL(default), variable name forxwill be used.- ylab
Labels for
yaxis variable. IfNULL(default), variable name forywill be used.- title
The text for the plot title.
- subtitle
The text for the plot subtitle. Will work only if
results.subtitle = FALSE.- caption
The text for the plot caption. This argument is relevant only if
bf.message = FALSE.- ggtheme
A
{ggplot2}theme. Default value istheme_ggstatsplot(). Any of the{ggplot2}themes (e.g.,ggplot2::theme_bw()), or themes from extension packages are allowed (e.g.,ggthemes::theme_fivethirtyeight(),hrbrthemes::theme_ipsum_ps(), etc.). But note that sometimes these themes will remove some of the details that{ggstatsplot}plots typically contains. For example, if relevant,ggbetweenstats()shows details about multiple comparison test as a label on the secondary Y-axis. Some themes (e.g.ggthemes::theme_fivethirtyeight()) will remove the secondary Y-axis and thus the details as well.- ggplot.component
A
ggplotcomponent to be added to the plot prepared by{ggstatsplot}. This argument is primarily helpful forgrouped_variants of all primary functions. Default isNULL. The argument should be entered as a{ggplot2}function or a list of{ggplot2}functions.- ...
Currently ignored.
Details
For details, see: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggscatterstats.html
Note
The plot uses ggrepel::geom_label_repel() to attempt to keep labels
from over-lapping to the largest degree possible. As a consequence plot
times will slow down massively (and the plot file will grow in size) if you
have a lot of labels that overlap.
Summary of graphics
| graphical element | geom used | argument for further modification |
| raw data | ggplot2::geom_point() | point.args |
| labels for raw data | ggrepel::geom_label_repel() | point.label.args |
| smooth line | ggplot2::geom_smooth() | smooth.line.args |
| marginal histograms | ggside::geom_xsidehistogram(), ggside::geom_ysidehistogram() | xsidehistogram.args, ysidehistogram.args |
Correlation analyses
The table below provides summary about:
statistical test carried out for inferential statistics
type of effect size estimate and a measure of uncertainty for this estimate
functions used internally to compute these details
Hypothesis testing and Effect size estimation
| Type | Test | CI available? | Function used |
| Parametric | Pearson's correlation coefficient | Yes | correlation::correlation() |
| Non-parametric | Spearman's rank correlation coefficient | Yes | correlation::correlation() |
| Robust | Winsorized Pearson's correlation coefficient | Yes | correlation::correlation() |
| Bayesian | Bayesian Pearson's correlation coefficient | Yes | correlation::correlation() |
Examples
set.seed(123)
# creating a plot
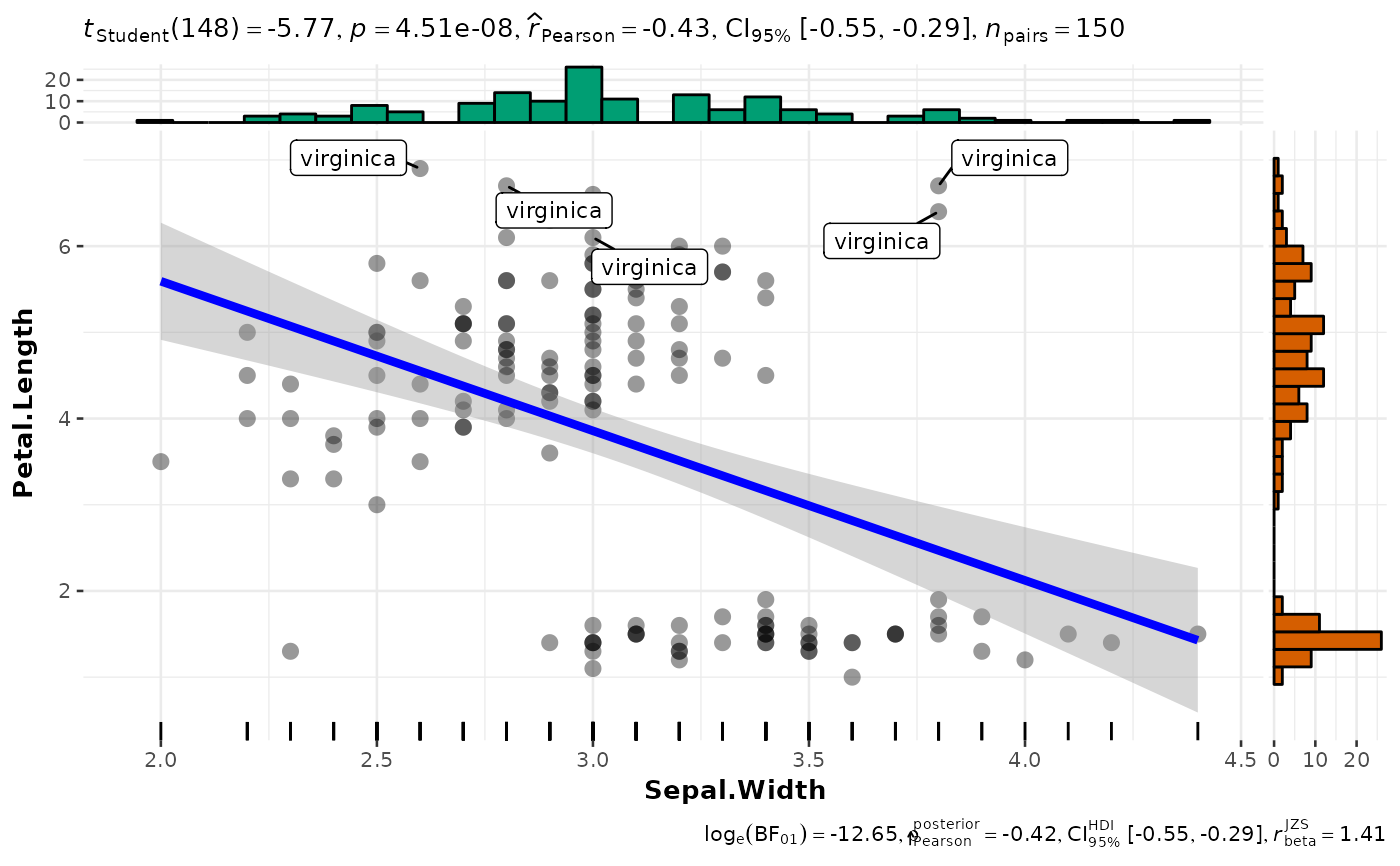
p <- ggscatterstats(
iris,
x = Sepal.Width,
y = Petal.Length,
label.var = Species,
label.expression = Sepal.Length > 7.6
) +
ggplot2::geom_rug(sides = "b")
# looking at the plot
p
#> `stat_xsidebin()` using `bins = 30`. Pick better value `binwidth`.
#> `stat_ysidebin()` using `bins = 30`. Pick better value `binwidth`.
 # extracting details from statistical tests
extract_stats(p)
#> $subtitle_data
#> # A tibble: 1 × 14
#> parameter1 parameter2 effectsize estimate conf.level conf.low
#> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Sepal.Width Petal.Length Pearson correlation -0.428 0.95 -0.551
#> conf.high statistic df.error p.value method n.obs
#> <dbl> <dbl> <int> <dbl> <chr> <int>
#> 1 -0.288 -5.77 148 0.0000000451 Pearson correlation 150
#> conf.method expression
#> <chr> <list>
#> 1 normal <language>
#>
#> $caption_data
#> # A tibble: 1 × 17
#> parameter1 parameter2 effectsize estimate conf.level
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 Sepal.Width Petal.Length Bayesian Pearson correlation -0.422 0.95
#> conf.low conf.high pd rope.percentage prior.distribution prior.location
#> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 -0.551 -0.290 1 0 beta 1.41
#> prior.scale bf10 method n.obs conf.method expression
#> <dbl> <dbl> <chr> <int> <chr> <list>
#> 1 1.41 312665. Bayesian Pearson correlation 150 HDI <language>
#>
#> $pairwise_comparisons_data
#> NULL
#>
#> $descriptive_data
#> NULL
#>
#> $one_sample_data
#> NULL
#>
#> $tidy_data
#> NULL
#>
#> $glance_data
#> NULL
#>
#> attr(,"class")
#> [1] "ggstatsplot_stats" "list"
# extracting details from statistical tests
extract_stats(p)
#> $subtitle_data
#> # A tibble: 1 × 14
#> parameter1 parameter2 effectsize estimate conf.level conf.low
#> <chr> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Sepal.Width Petal.Length Pearson correlation -0.428 0.95 -0.551
#> conf.high statistic df.error p.value method n.obs
#> <dbl> <dbl> <int> <dbl> <chr> <int>
#> 1 -0.288 -5.77 148 0.0000000451 Pearson correlation 150
#> conf.method expression
#> <chr> <list>
#> 1 normal <language>
#>
#> $caption_data
#> # A tibble: 1 × 17
#> parameter1 parameter2 effectsize estimate conf.level
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 Sepal.Width Petal.Length Bayesian Pearson correlation -0.422 0.95
#> conf.low conf.high pd rope.percentage prior.distribution prior.location
#> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 -0.551 -0.290 1 0 beta 1.41
#> prior.scale bf10 method n.obs conf.method expression
#> <dbl> <dbl> <chr> <int> <chr> <list>
#> 1 1.41 312665. Bayesian Pearson correlation 150 HDI <language>
#>
#> $pairwise_comparisons_data
#> NULL
#>
#> $descriptive_data
#> NULL
#>
#> $one_sample_data
#> NULL
#>
#> $tidy_data
#> NULL
#>
#> $glance_data
#> NULL
#>
#> attr(,"class")
#> [1] "ggstatsplot_stats" "list"